The EPS Database provides access to detailed structural, taxonomic and bibliographic information on bacterial EPS. Each EPS entry in the database is represented the repeating unit structure and linked to various EPS structures expressed by different strains of the same bacterium. Bacterial source and growth conditions for EPS expression and structure determination are likewise listed when available. To provide access to further structural information such as the WURCS and GlycoCT code the EPSs are linked to the GlyTouCan international structure repository. Literature links are likewise provided.
The EPS Database is based on retrospective literature analysis and links to original structure analysis are provided. The EPS data are accessible in its entirety with several ordering functionalities or by making use of one or more search filters including organism, strain, backbone and branch composition, author and publication data.
The EPS Database is primarily expanded by the depositing functionality where authors are encouraged to deposit the elucidated EPS structure which will be added to the database as the structure is accepted for publication. Each deposited EPS structure is checked manually. The attempt is to cover all bacterial EPS structures across bacterial organisms so please use the deposit or contact function if we missed some. The EPS Database is available at no cost and requires no registration.
Latest entry
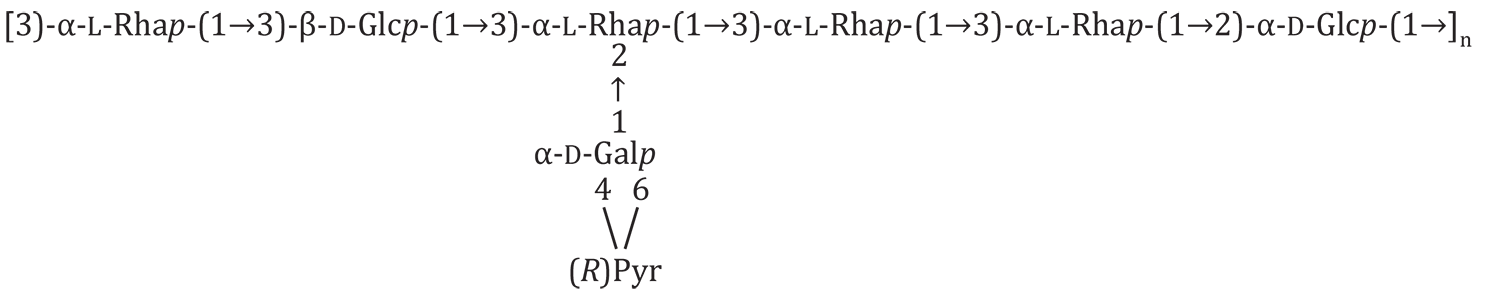
Uploaded by Marie-Rose Van Calsteren, June 2016
Reference